Using Human Cells for Broadly Reactive Influenza Antibody Discovery
The Challenge
- Isolating antigen-specific, antibody-secreting cells from human donor samples
- Screen for antibodies with rare activity using multilayer screening plan
The Single Cell Solution
- Adapt AbTheneum to work with human cells
- Build a customized workflow for enriching antigen-specific antibodies
- Successfully find rare events by building a multi-parameter screening plan
Highlights
- Isolating antigen-specific, antibody-secreting cells from human donor samples.
- Screen for antibodies with rare activity using multilayer screening plan.
Challenge: Demonstrate Key Capabilities of The AbTheneumTM Platform
These include:- A workflow using human cells instead of mouse
- A customized workflow for enriching antigen-specific antibodies using a cell sorter
- The ability to “find a needle in a haystack” – in this case, hunting for the rare antibodies that react with 4 very different molecules
This case study was made in collaboration with Miltenyi Biotec.
Solution: Finding a Better Way
Cell Isolation
AbTheneum™ uses antibody-secreting cells. For mice, there are commercially available kits to isolate plasma cells, and they work pretty well. For human cells, there are many other commercially available kits, which we used some in combination with other in-house methods to get to our goal – antibody-secreting cells from humans that we can enrich for target-specific ones.
Our collaborator, Miltenyi Biotec, sourced peripheral mononuclear blood cells (PBMCs) from 3 donors that had received their seasonal influenza vaccine within 6 weeks. We thawed the PBMCs several days before the scheduled AbTheneum screen and isolated Class-Switched Memory B Cells (CSMBCs) using a kit by Miltenyi (catalog # 130-093-617). The kit yields “untouched” cells, which is perfect for this workflow as we want to apply positive selection on the CSMBCs to enrich for anti-influenza antibody-secreting cells. The CSMBCs have antibodies expressed on the surface but are not secreting. These cells can be activated to secrete using different cytokines and cofactors, similar to the B Cell Expansion Kit available from Miltenyi (catalog # 130-106-196). Single Cell used an in-house cytokine cocktail to activate cells over 5 days. On Day 3, cells were enriched for target-specific cells using the MACSQuant Tyto cell sorter.
On Day 5, the activated memory B cells were enriched by MACSQuant Tyto by staining cells with fluorescently-labeled anti-CD38, 7-AAD, and 4 influenza HA proteins. Cells were sorted for CD38+/7-AAD-/HA+ to isolate antibody-secreting, antigen-specific cells. Fig 1 shows the sort gate and frequency of events in this cell isolation.
Figure 1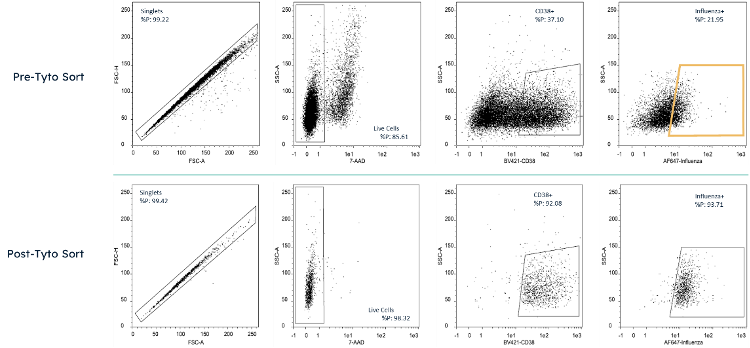 Cell analysis plot taken by MACSQuant 10 of cells pre and post sort by MACSQuant Tyto. Cells were stained with 7-AAD, Anti-CD38 Brilliant Violet 421, and 4 influenza HA proteins in AlexaFluor 647. The sort gate for Tyto sorting is highlighted in yellow.
Cell analysis plot taken by MACSQuant 10 of cells pre and post sort by MACSQuant Tyto. Cells were stained with 7-AAD, Anti-CD38 Brilliant Violet 421, and 4 influenza HA proteins in AlexaFluor 647. The sort gate for Tyto sorting is highlighted in yellow.
We selected the 4 HA proteins (Table 1) used for cell isolation because they cover the 2 groups within the influenza A family and are therefore highly diverse compared to each other. Our goal is to mine the antibody repertoire from these donor samples to find antibodies that have ability to bind to diverse strains of influenza protein, in order to be useful in a pandemic influenza event.
Table 1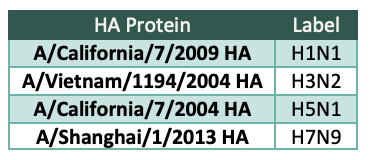 HA proteins used for screening and the label used for these HA proteins in this document.
HA proteins used for screening and the label used for these HA proteins in this document.
AbTheneum Screening
Cells sorted from Tyto were deposited onto picoliter devices. Antibodies secreted from the cells were captured onto 3 capture slides and screened according to the Screening Plan (Fig 2). The Screening Plan was designed to allow assigning antibodies to categories (e.g. highly specific to a single HA, or cross-reactive to 2, 3, or 4 HAs). The Screening Plan also includes assessing all captured antibodies for their ability to compete with a benchmark antibody, MEDI8852, which is currently being evaluated in clinical trials as a broadly-neutralizing anti-influenza therapy. After staining each slide with the stain condition in the Screening Plan, the slides are scanned in a fluorescent scanner and images are generated. This project has sequential stains on 3 slides, generating 8 images for each picoliter device. A small region of the images is cropped and shown for all the stain conditions in Fig 3.
Figure 2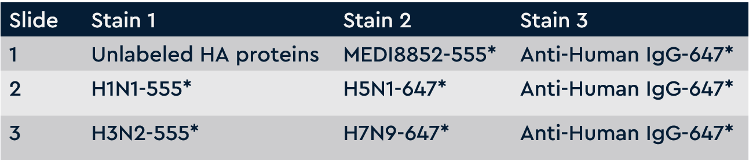 Screening plan for broadly-cross subtype reactive anti-influenza antibodies. Each slide is washed and scanned on a fluorescent slide scanner after each stain. Slide 1 screens all antibodies for competition with MEDI8852 antibody (negative hits are competing, positive hits are non-competing). Slide 2 screens all antibodies for binding to H1N1 and H5N1. Slide 3 screens all antibodies for binding to H3N2 and H7N9. All slides detect all IgGs on the last stain.
Screening plan for broadly-cross subtype reactive anti-influenza antibodies. Each slide is washed and scanned on a fluorescent slide scanner after each stain. Slide 1 screens all antibodies for competition with MEDI8852 antibody (negative hits are competing, positive hits are non-competing). Slide 2 screens all antibodies for binding to H1N1 and H5N1. Slide 3 screens all antibodies for binding to H3N2 and H7N9. All slides detect all IgGs on the last stain.
Figure 3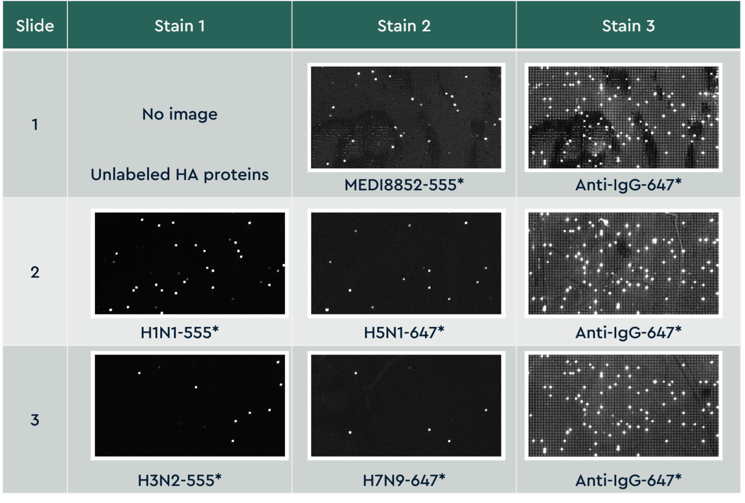 Small cropped region from the same area on all 3 slides across 8 staining conditions. Slides were imaged on a fluorescent slide scanner that has 2 lasers capable of detecting dyes excited at 532 and 635 nm. Images are aligned across all slides and images, analyzed for fluorescent intensity, and individual antibody binding properties are reported for each screen condition.
Small cropped region from the same area on all 3 slides across 8 staining conditions. Slides were imaged on a fluorescent slide scanner that has 2 lasers capable of detecting dyes excited at 532 and 635 nm. Images are aligned across all slides and images, analyzed for fluorescent intensity, and individual antibody binding properties are reported for each screen condition.
After antibody capture and profiling, antibodies were sequenced in parallel by lysing all cells, capturing encoding mRNA, and sequencing by NGS. The output from 2,593 antigen-specific antibodies profiled is displayed in the heat map chart (Fig 4).
Figure 4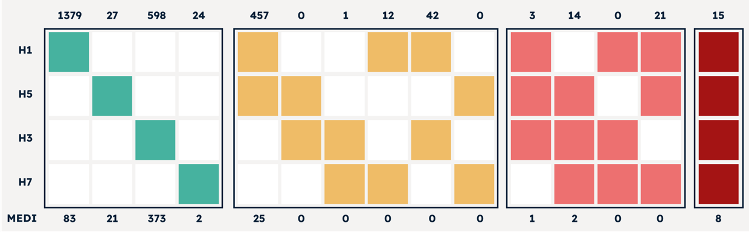 Data output of 2,593 antibodies discovered by AbTheneum. Each antibody was screened for binding against the H1N1, H5N1, H3N2, and H7N9 HA proteins. All 15 possible binding properties are shown using colored columns. For example, H1 and H5 colored yellow with H3 and H7 white means antibodies that bind to H1N1 and H5N1, with no binding to H3N2 and H7N9. The top x-axis lists the count of mAbs that exhibit the binding profile each colored column. The bottom x-axis lists the number of mAbs in the group that compete with MEDI8852.
Data output of 2,593 antibodies discovered by AbTheneum. Each antibody was screened for binding against the H1N1, H5N1, H3N2, and H7N9 HA proteins. All 15 possible binding properties are shown using colored columns. For example, H1 and H5 colored yellow with H3 and H7 white means antibodies that bind to H1N1 and H5N1, with no binding to H3N2 and H7N9. The top x-axis lists the count of mAbs that exhibit the binding profile each colored column. The bottom x-axis lists the number of mAbs in the group that compete with MEDI8852.
After recovering sequences from the screened antibodies, Single Cell selected 5 antibodies with varied binding profiles as a first round to validate the screening result. ELISA against the 4 HA proteins confirmed the SCT screening results for all 5 antibodies (Fig 5).
Figure 5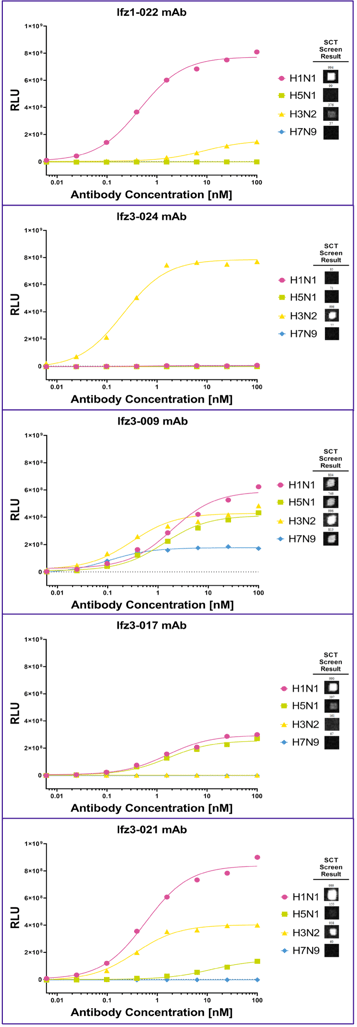 Five expressed antibodies (Ifz1-022, Ifz3-24, Ifz3-009, Ifz3-017, and Ifz3-021) were evaluated by ELISA for their binding against different HA proteins. The SCT Screen Result is shown as image cutouts to each of the HA proteins after screening according to the screening plan.
Five expressed antibodies (Ifz1-022, Ifz3-24, Ifz3-009, Ifz3-017, and Ifz3-021) were evaluated by ELISA for their binding against different HA proteins. The SCT Screen Result is shown as image cutouts to each of the HA proteins after screening according to the screening plan.
Outcome: Moving the Campaign Ahead
This case study demonstrates 3 different abilities that AbTheneum can address:
- Adapting AbTheneum to work with human cells, including adapting primers for sequencing and building a cell isolation workflow addressing human cells instead of mouse
- Building a customized workflow for enriching antigen-specific antibodies, enabled by the MACSQuant Tyto cell sorter
- A successful quest to seek rare events by building a multi-parameter screening plan