Upfront Blocking Assay to Discover Neutralizing Anti-SARS-CoV-2 Antibodies
The Challenge
- Screening for antibodies that block the interaction between ligand and receptor
- Validating blocking with pseudovirus neutralization assays
The Single Cell Solution
- Develop a blocking assay that has a high correlation with neutralizing the virus
- Set up the assay in-line during discovery to save downstream time
Highlights
- Screening for antibodies that block the interaction between ligand and receptor
- Validated blocking with pseudovirus neutralization assays
Problem: Taking on the Challenge
SARS-CoV-2 infects cells by the RBD on the S1 subunit of the spike protein binds with human ACE2 receptor (Figure 1). Many therapeutic efforts to target SARS-CoV-2 have focused on blocking this interaction.
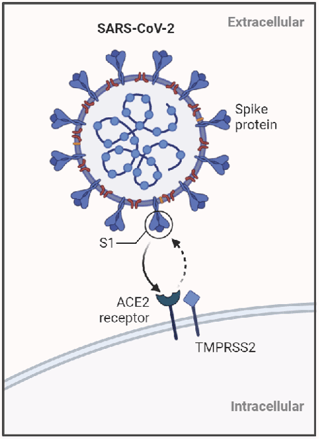 Figure 1
Figure 1
In fact, many therapeutic targets have the same goal – block the interaction between the ligand and receptor.
Solution: Finding a Better Way
Using the SARS-CoV-2 pandemic as a model case study, Single Cell developed a blocking assay in-line to AbTheneum™ that had a high correlation with neutralizing the virus.
We immunized a cohort of wild-type CD-1 mice with recombinant RBD protein. The antibody-secreting cells were isolated using a CD138+ isolation kit. The antibodies were screened by AbTheneum, depositing cells into our device, generating antibody microarrays, and screened those microarrays. Figure 2 shows a schematic of the generation of antibody microarrays from antibody-secreting cells as part of the AbTheneum workflow. The microarrays allows all antibodies to be assayed simultaneously. For this work, we developed a novel blocking assay to assess each antibody’s ability to block the interaction of the spike protein and human ACE2 receptor.
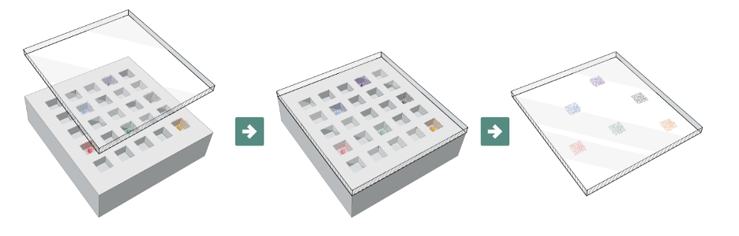 Figure 2
Figure 2
The blocking assay is illustrated in Figure 3. Two antibody microarrays were generated, called Slide 1 and Slide 2. Slide 1 identifies all secreted antibodies that bind recombinant fluorescently-labeled RBD and also allow recombinant fluorescently-labeled ACE2 to bind (non-blockers). Slide 2 identifies all secreted antibodies that bind to recombinant RBD (all binders). All images are aligned, and blocking antibodies are identified by observing positive signal on Slide 2 and negative signal on Slide 1. We define these antibodies as “blocking” antibodies.
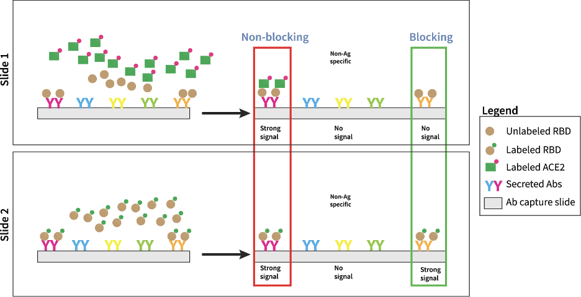 Figure 3
Figure 3
After generating antibody microarrays, all cells are lysed and mRNA captured and sequenced en masse as part of the standard AbTheneum workflow. All IgGs are sequenced by Next Generation Sequencing, and AbTheneum informatics aligns the antibody microarray data with the full-length sequence pairs recovered from NGS.
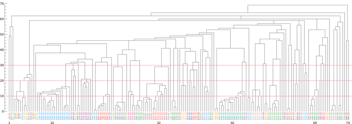
Over 1900 RBD-specific antibody sequences were recovered and clustered by sequence similarity. We analyzed the sequences from the discovered antibodies to compare families of similar sequences and their activity in the blocking assay. We found that similar sequence families (10 amino acid changes or less across full-length VH-VL) showed similar activity in the blocking assay. Figure 4 shows related clusters with each antibody’s signal displayed from Slide 1 and Slide 2.
17 antibodies with high sequence diversity were selected for recombinant expression. 2 antibodies did not express. The remaining 15 had 13 show blocking activity in our blocking assay (labeled in pink in Fig 4), with 2 non-blocking (labeled in blue in Fig 4), included to show the predictive power of the blocking assay. Expressed antibodies were tested using a SARS-CoV-2 reporter virus particles (RVPs) by Integral Molecular. The RVPs have a modified genome that expresses luciferase upon cellular infection. Luciferase was quantified by using Renilla-GLO system by Promega on the GloMax Navigator. We define antibodies that inhibit RVPs as “neutralizing” antibodies, not to be confused with “blocking” antibodies as defined previously. The RVPs had the widespread D614G mutation in the pseudovirus construct. Published antibodies CA1 and CB6 recovered from convalescent patients (Shi et al.) were reconstructed and run as positive controls alongside Single Cell’s antibodies.
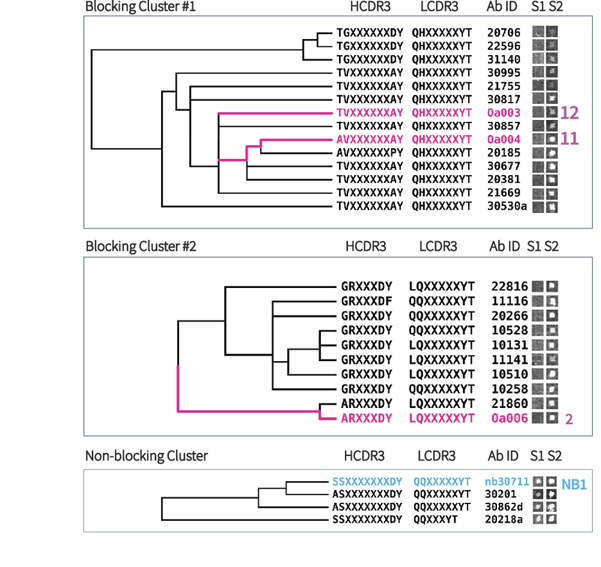 Figure 4
Figure 4
Figure 5 shows the results from the pseudovirus neutralization assay. 8 out of 15 blocking antibodies performed better than published controls. The two non-blocking antibodies (NBAb 1 and NBAb2) showed no and low potency in the assay.
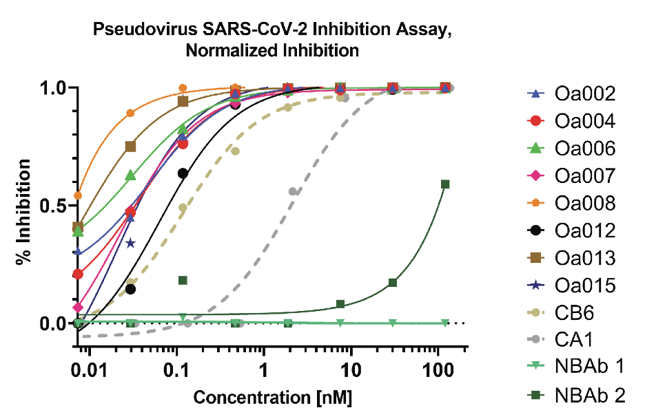 Figure 5
Figure 5
Outcome: Moving the Campaign Ahead
The blocking assay is useful for other targets where a recombinant receptor can be synthesized. The blocking assay is performed in-line during discovery, which saves downstream time.
References:
Shi, R., Shan, C., Duan, X. et al. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature 584, 120-124 (2020).

-1.png?width=352&name=freezer%20boxes%20in%20-80C%20(1)-1.png)